
Written by Emanuele Casali, Stefano A. Serapian, Eleonora Gianquinto, Matteo Castelli, Massimo Bertinaria, Francesca Spyrakis, Giorgio Colombo
Recent research has suggested that targeting the NLRP3 inflammasome may be a promising strategy for easing the symptoms of neuroinflammation associated with several pathologic conditions such as long COVID-19, Alzheimer’s disease, and Parkinson Disease. The central core of the inflammasome complex is the NLRP3 protein. When the inflammasome is activated, NLRP3 undergoes a significant conformational change that triggers the production of pro-inflammatory cytokines IL-1β and IL-18, as well as cell death by pyroptosis. Small molecules binding to an allosteric site proved able to induce NLRP3 inhibition, but the usage in clinics of known inhibitors has been hampered by their toxicity. In this context, the necessity of inhibitors with a better pharmacological profile remains an open issue. To meet this goal, it is crucial to complement the available structural data with approaches that permit to unveil the impact of ligands on the functionally oriented dynamics of NLRP3.
Thanks to the resources provided by Fenix infrastructure, a joint team from the University of Pavia and the University of Turin has investigated the origins of allosteric inhibition of NLRP3. Through molecular dynamics (MD) simulations and advanced analysis methods, researchers have provided molecular-level insights into how binding of allosteric inhibitors affects protein structure and dynamics, with key reverberations on how NLRP3 is preorganized for assembly and ultimately function. The results revealed ligand-dependent structural remodelling of the conformational ensembles populated by the protein, a modification of the motion of functional domains, and a reorganization of the substructures dedicated to engaging other molecular partners.
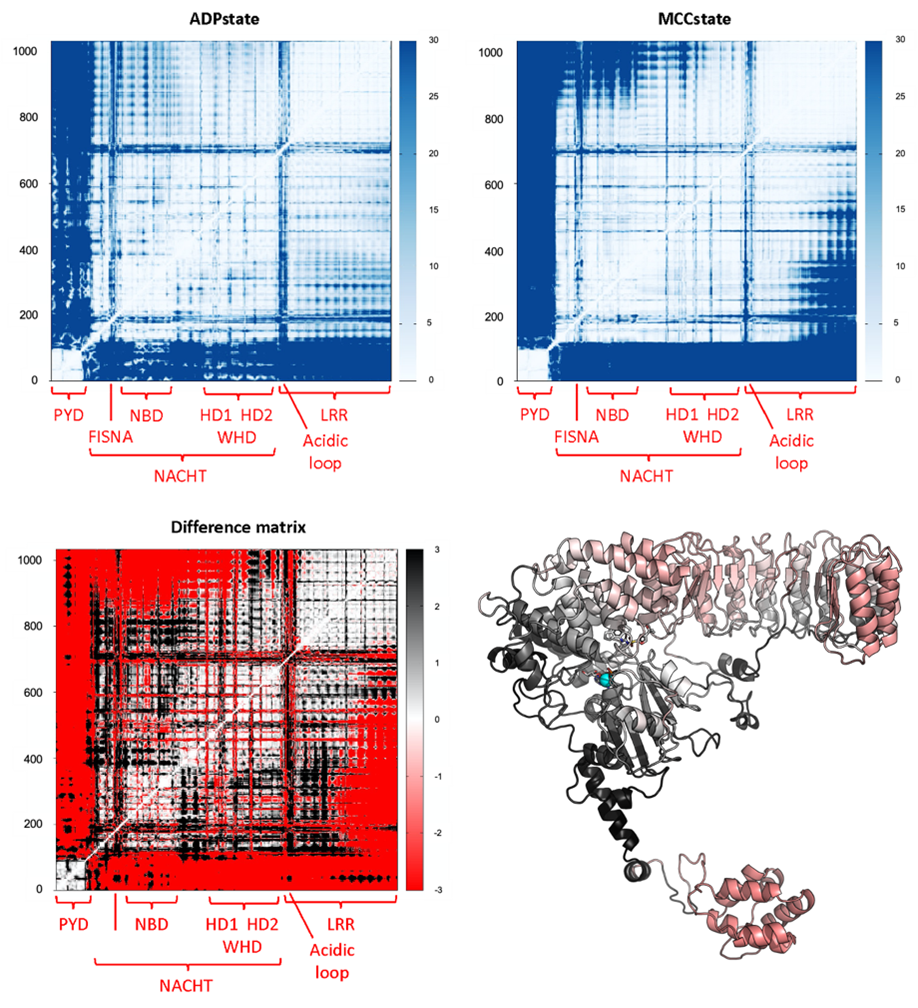
Figure 1. Distance fluctuation (DF) matrices (in the absence and presence of the allosteric inhibitor MCC); left and right respectively) and difference matrix. Redder areas denote a loss of coordination upon adding MCC, whereas blacker areas denote a loss of coordination upon removing MCC.
In this framework, a mechanistic model was developed to reconnect the inhibitor-induced effects on the protein’s microscopic dynamics to the observed (macroscopic) inactivation of NLRP3. A new and efficient Machine Learning (ML) protocol was then developed to analyse protein dynamics data and discern the dynamic signature differentiating active vs. inactive/inhibited states.
This method allows classifying designed ligands as hits (those that induce inactive protein dynamic traits) vs. non-hits (those that do not alter the dynamics of the protein active state). The knowledge generated herein is currently being used to guide the design of novel allosteric ligands targeting NLRP3, thus expanding the chemical space of NLRP3 inhibitors and the range of possible interventions.
E. Casali, S.A. Serapian, E. Gianquinto, M. Castelli, M. Bertinaria, F. Spyrakis and G. Colombo. “NLRP3 monomer functional dynamics: from the effects of allosteric binding to implications for drug design” Int. J. Biol. Macromol. 246:125609, 2023. 10.1016/j.ijbiomac.2023.125609
Acknowledgements
We acknowledge the use of Fenix Infrastructure resources, which are partially funded by the European Union’s Horizon 2020 research and innovation programme through the ICEI project under the grant agreement No. 800858. This study was further supported by the European Union’s Horizon 2020 Framework Programme for Research and Innovation under Specific Grant Agreements No. 945539 (HBP SGA3).